Under the Hood: A New Engine for Modeling Biological Processes
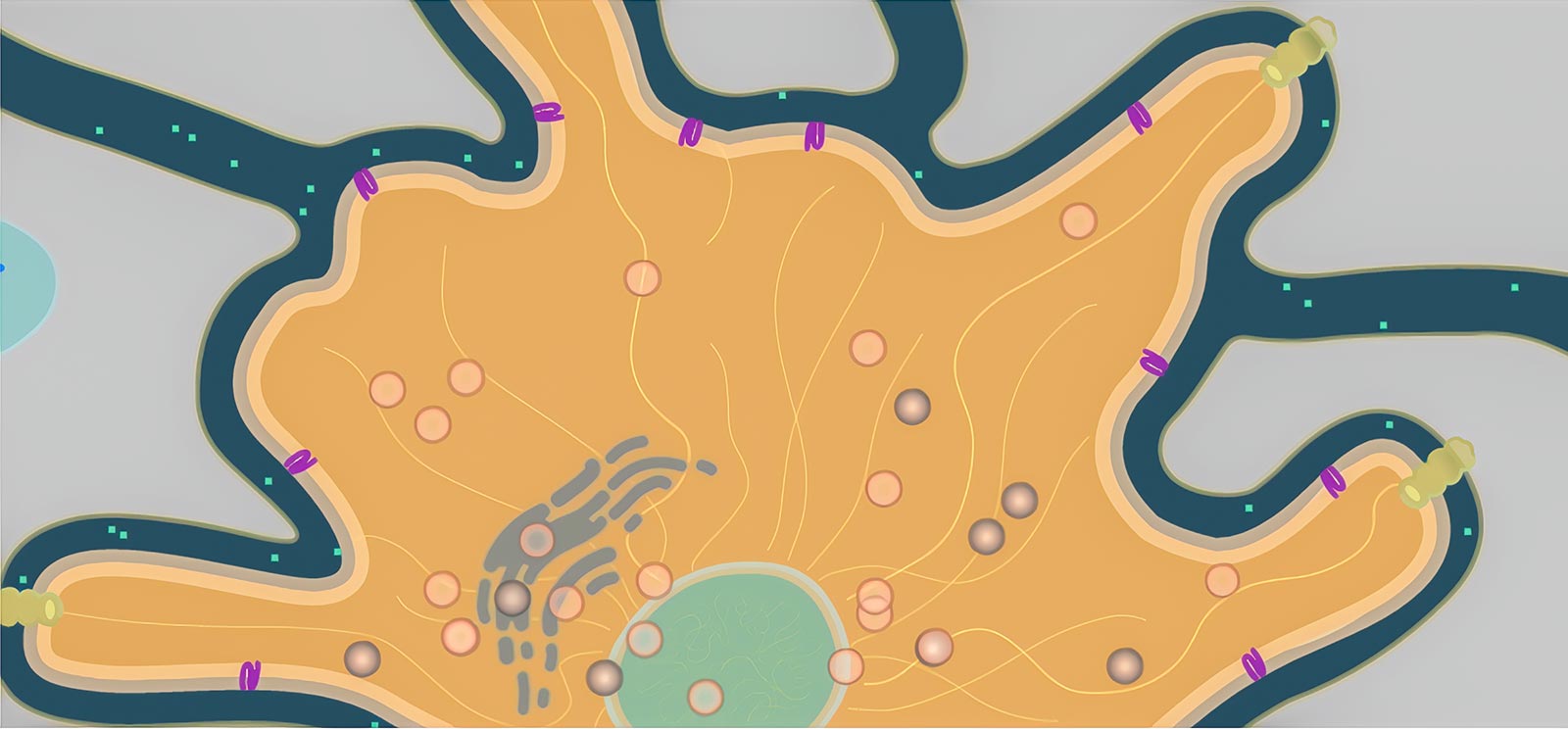
Consider a tiny hormone as it flows between cells and binds to a receptor protein in a cell’s membrane. The protein, if working correctly, triggers a series of reactions that releases a signal molecule headed for the cell’s nucleus, which in response produces a strand of mRNA. This mRNA finds its way to the endoplasmic reticulum, where it causes melanin to be packed into a tiny organelle called a melanosome. The little bundle of melanin gets carried through the cell by protein motors, where it is anchored in the best location to absorb light and darken the cell. All this happens hundreds of times a second throughout this cell and all of its neighbors.
To illustrate such cellular processes for our Geniventure dragon genetics game, we needed to develop a new modeling engine. After deciding not to attempt the impossible task of modeling a cell from physics first principles (like we do with Molecular Workbench), a rule-based agent model seemed like a good solution. However, while a number of agent-based modeling engines exist, from NetLogo to our own Populations engine (which we use to model ecosystems and evolution), none of them seemed to fit the bill. We wanted to make models that looked organic and beautiful, that could be created fairly easily even by non-programmers, and that could be reused to make many different cells without needing to rewrite the same rules each time.
We created a new engine called Organelle with two unique features (Figure 1). First, model environments are created using SVG, a vector-based image format that allows us to draw features of the environment and name them semantically. That is, we can label the parts (Golgi apparatus, microtubule, receptor protein) and refer to them directly in the rules for the agents (“move towards the nearest receptor protein”). Second, we can define agents—the individual moving bodies in the cell, such as proteins and vesicles—and give them rules that can be reused in any other model. So we only have to define, say, a melanosome (the organelle that packages melanin) once, and it will behave the same way in other models, though the presence of new features or other agents may cause it to behave differently.
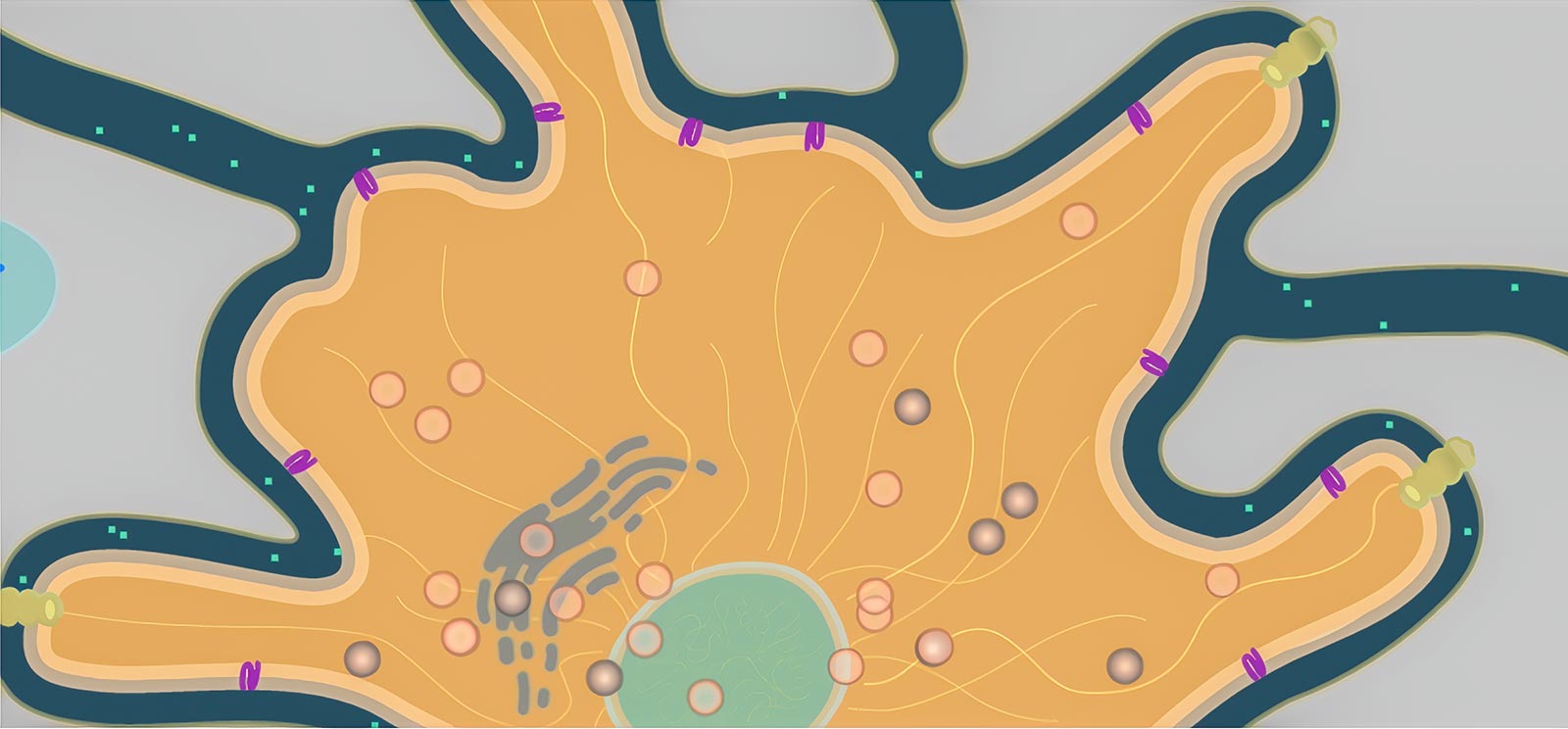
To create the rules for these agents, an author describes their behavior using verbs that are meaningful for the kinds of models we are creating: “grow,” “flow,” “diffuse,” “follow,” “find,” etc. The aim is to create a readable set of instructions that can be understood even by a non-programmer. (These verbs can be expanded by a plug-in system to allow programmers to create all kinds of behaviors.)
A sample set of rules written in YAML can be seen in Figure 2. The code instructs the model to spawn a new “hormone” agent every 20 model ticks at one of the SVG elements named “intercellular-path,” if any exist in the current model. Once spawned, this agent will simply follow that path until the end, where it will be removed from the model. Naturally these rules can become much more complex.
Our open-source Organelle engine is also used in our Connected Biology project, which links genetics and evolution, but it can be used for more than cellular models—the engine can work anywhere agent-based models can benefit from semantically defined environments and simple, portable rules. Find demos of these models and try modifying them by editing the rules directly in the browser at the Organelle homepage.
name: hormone image: assets/hexagon.svg properties: speed: 1.2 spawn: every: 20 at: selector: "#intercellular-path" which: random initialState: flowing rules: flowing: - follow: selector: "#intercellular-path" which: nearest finally: die: true
Sam Fentress (sfentress@concord.org) is a senior software engineer.
This material is based upon work supported by the National Science Foundation under grant DRL-1620910. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.